Parsing data from the Sonardyne FETCH AZA
The purpose of this notebook is to demonstrate the functionality of fetchAZA python package.
The demo is organised to show
Step 1: Reading the *.csv files into xarray datasets
Step 2: Writing the xarray datasets into individual netCDF files
Step 3: Various plots
Note that when you submit a pull request, you should clear all outputs from your python notebook for a cleaner merge.
[1]:
import pathlib
import sys
script_dir = pathlib.Path().parent.absolute()
parent_dir = script_dir.parents[0]
sys.path.append(str(parent_dir))
import xarray as xr
import os
import numpy as np
import matplotlib.pyplot as plt
import importlib
import datetime
from fetchAZA import convertAZA, readers, writers, plotters, tools, timetools, utilities
import warnings
import re
import glob
import logging
_log = logging.getLogger(__name__)
# Specify the path for writing datafiles
data_path = os.path.join(parent_dir, 'data')
fig_path = os.path.join(parent_dir, 'figures')
warnings.filterwarnings("ignore", message="In a future version of xarray decode_timedelta will default to False rather than None. To silence this warning, set decode_timedelta to True, False, or a 'CFTimedeltaCoder' instance.")
warnings.filterwarnings("ignore", category=xr.SerializationWarning, message="SerializationWarning: Can't decode floating point timedelta to 's' without precision loss, decoding to 'ns' instead. To silence this warning use time_unit='ns' in call to decoding function.")
Step 1 & 2 as convertAZA.convertAZA
[2]:
fn = 'sample_data.csv'
STN = 'sample'
deploy_date = '2023-02-27'
recovery_date = '2023-03-08T08:00:00'
latitude = 26.5
longitude = -76.75
water_depth = -3800
print("Calling convertAZA with overwrite=True...")
print("This should NOT prompt for overwrite confirmation.")
# Force reload the module to pick up any changes
import importlib
importlib.reload(convertAZA)
# Test the overwrite parameter directly first
print(f"convertAZA function defaults: {convertAZA.convertAZA.__defaults__}")
ds_pressure, ds_AZA = convertAZA.convertAZA(
data_path,
fn,
STN,
deploy_date,
recovery_date,
latitude,
longitude,
water_depth,
cleanup=True,
overwrite=True
)
print("Function completed successfully!")
Calling convertAZA with overwrite=True...
This should NOT prompt for overwrite confirmation.
convertAZA function defaults: ('sample', '2000-01-01', '2099-01-01', '0', '0', '0', ['DQZ', 'PIES', 'INC', 'TMP', 'KLR'], True, None)
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for join will change from join='outer' to join='exact'. This change will result in the following ValueError: cannot be aligned with join='exact' because index/labels/sizes are not equal along these coordinates (dimensions): 'new_index' ('new_index',) The recommendation is to set join explicitly for this case.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/data/sample_data*.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_KLR.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_DQZ.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_AZAseq.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_PIES.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_TMP.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_INC.nc
Dataset AZAseq not included in combined datasets
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/tools.py:353: FutureWarning: In a future version of xarray the default value for join will change from join='outer' to join='exact'. This change will result in the following ValueError: cannot be aligned with join='exact' because index/labels/sizes are not equal along these coordinates (dimensions): 'RECORD_TIME' ('RECORD_TIME',) The recommendation is to set join explicitly for this case.
combined_dataset = xr.merge(combined_datasets.values(), compat="override")
Deleting file: /home/runner/work/fetchAZA/fetchAZA/data/sample_data_KLR.nc
Deleting file: /home/runner/work/fetchAZA/fetchAZA/data/sample_data_DQZ.nc
Deleting file: /home/runner/work/fetchAZA/fetchAZA/data/sample_data_PIES.nc
Deleting file: /home/runner/work/fetchAZA/fetchAZA/data/sample_data_TMP.nc
Deleting file: /home/runner/work/fetchAZA/fetchAZA/data/sample_data_INC.nc
Function completed successfully!
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/writers.py:129: UserWarning: Times can't be serialized faithfully to int64 with requested units 'seconds since 1970-01-01'. Resolution of 'milliseconds' needed. Serializing times to floating point instead. Set encoding['dtype'] to integer dtype to serialize to int64. Set encoding['dtype'] to floating point dtype to silence this warning.
ds.to_netcdf(output_file)
Step 1: Read the *csv file for Logging Events.
This is done with readers.process_csv_to_xarray(). All logging events are read into individual xarray datasets, stored as a dictionary of datasets where the key. In addition, the AZA sequence (events following the pattern AZS-AZA-AZA-AZA-AZS) are read into an additional dataset with key ‘AZAseq’. Since this dataset does not contain every individual AZA or AZS event, it does not replace the individual datasets.
A log of the processing is also generated.
Optionally, the deployment and recovery dates can be passed. If they are, then the datasets will be sliced to these dates.
[3]:
fn = 'sample_data.csv'
STN = 'sample'
deploy_date = '2023-02-27'
recovery_date = '2023-03-08T08:00:00'
# Process filename
file_path = os.path.join(data_path, fn)
file_root = fn.split('.')[0]
platform_id = file_root
today = datetime.datetime.now()
start_time = today.strftime("%Y%m%dT%H")
# Create a log file
log_file = os.path.join(data_path, f"{platform_id}_{start_time}_read.log")
logf_with_path = os.path.join(data_path, log_file)
logging.basicConfig(
filename=logf_with_path,
encoding='utf-8',
format="%(asctime)s %(levelname)-8s %(funcName)s %(message)s",
filemode="w", # 'w' to overwrite, 'a' to append
level=logging.INFO,
datefmt="%Y%m%dT%H%M%S",
force=True,
)
_log.info('Reading AZA from CSV to netCDF')
_log.info('Processing data from: %s', file_path)
# Process the CSV file and create xarray datasets containing the data
datasets = readers.read_csv_to_xarray(file_path)
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for join will change from join='outer' to join='exact'. This change will result in the following ValueError: cannot be aligned with join='exact' because index/labels/sizes are not equal along these coordinates (dimensions): 'new_index' ('new_index',) The recommendation is to set join explicitly for this case.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/readers.py:375: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
combined_dataset = xr.merge(list(datasets.values()))
Step 2: Write the data to netCDF
[4]:
writers.save_datasets(datasets, file_path, overwrite=True)
Step 3: Further processing of pressure records and AZA sequence records
Note that in the steps above, the original data were not changed, with the exception of changes noted in the log file.
This means that each of the newly created *.nc mirrors–almost exactly–the original data.
Here we carry out additional steps including:
Load netCDF datasets based on provided keys. (
readers.load_netcdf_datasets(data_path, file_root, keys))Convert units and adjust time formats. (
timetools.convert_seconds_to_float(ds))Assign sampling time for the AZA sequence dataset. (
timetools.assign_sample_time())Filter datasets to the deployment period. (
timetools.cut_to_deployment(datasets, deploy_date, recovery_date))Reindex datasets on time. (
timetools.reindex_on_time(ds))Rename variables in datasets using predefined mappings. (using
vars_to_rename, a dict)Add dataset-specific attributes to variables.
Combine selected datasets into a single dataset. (using
xr.merge())Interpolate the combined dataset to an evenly spaced time grid. (after determining median interval of hourly with
timetools.calculate_sample_rate(ds))Clean and organize dataset attributes and variables.
Process the AZA sequence dataset, including renaming attributes and cleaning variables.
[5]:
ds_pressure, ds_AZA = tools.process_datasets(data_path, file_root, deploy_date, recovery_date)
ds_pressure
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/tools.py:353: FutureWarning: In a future version of xarray the default value for join will change from join='outer' to join='exact'. This change will result in the following ValueError: cannot be aligned with join='exact' because index/labels/sizes are not equal along these coordinates (dimensions): 'RECORD_TIME' ('RECORD_TIME',) The recommendation is to set join explicitly for this case.
combined_dataset = xr.merge(combined_datasets.values(), compat="override")
/home/runner/work/fetchAZA/fetchAZA/data/sample_data*.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_KLR.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_DQZ.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_AZAseq.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_PIES.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_TMP.nc
/home/runner/work/fetchAZA/fetchAZA/data/sample_data_INC.nc
Dataset AZAseq not included in combined datasets
[5]:
<xarray.Dataset> Size: 7kB
Dimensions: (TIME: 103)
Coordinates:
* TIME (TIME) datetime64[ns] 824B 2023-03-03T23:00:00 ... ...
Data variables: (12/14)
TIME_OF_FLIGHT_RMLE (TIME) float64 824B 5.11 5.11 5.11 ... 5.148 5.146
TIME_OF_FLIGHT_XCORR (TIME) float64 824B 5.11 5.11 5.109 ... 5.109 5.109
HALFLIFE (TIME) float32 412B 251.4 230.3 229.1 ... 345.3 335.9
MAGNITUDE (TIME) float32 412B 6.903e+03 9.485e+03 ... 1.506e+03
PEAK_POSITION (TIME) float32 412B 0.008512 0.008704 ... 0.007808
PITCH (TIME) float32 412B -1.875 -1.862 ... -1.853 -1.847
... ...
PRETRIGGER_NOISE (TIME) float32 412B 82.88 82.62 86.51 ... 64.36 65.5
RDIFF (TIME) float32 412B 178.6 183.0 169.2 ... 86.83 89.4
ROLL (TIME) float32 412B -0.1813 -0.1906 ... -0.1844
TEMPERATURE (TIME) float32 412B 3.073 2.9 2.756 ... 2.372 2.378
TEMPERATURE_DQZ (TIME) float32 412B 5.63 4.87 4.27 ... 2.43 2.43 2.43
TEMPERATURE_KLR (TIME) float32 412B 7.012 5.885 4.936 ... 2.215 2.214
Attributes:
Calculation_Version_PIES: 3
Index_DQZ: 3
Index_INC: 1
Index_KLR: 5
Index_PIES: 3
Index_TMP: 4
Serial Number: 1262636
Serial_Number_DQZ: 155399
Serial_Number_INC: 0
Serial_Number_KLR: 1262636
Serial_Number_TMP: 0
UID: 007217Save the data
[6]:
# Save the datasets
output_file = os.path.join(data_path, f"{STN}_{deploy_date.replace('-','')}_use.nc")
writers.save_dataset(ds_pressure, output_file, overwrite=True)
output_file = os.path.join(data_path, f"{STN}_{deploy_date.replace('-','')}_AZA.nc")
writers.save_dataset(ds_AZA, output_file, overwrite=True)
/home/runner/work/fetchAZA/fetchAZA/fetchAZA/writers.py:129: UserWarning: Times can't be serialized faithfully to int64 with requested units 'seconds since 1970-01-01'. Resolution of 'milliseconds' needed. Serializing times to floating point instead. Set encoding['dtype'] to integer dtype to serialize to int64. Set encoding['dtype'] to floating point dtype to silence this warning.
ds.to_netcdf(output_file)
[6]:
True
Plot variables in ds_pressure
[7]:
# Example usage
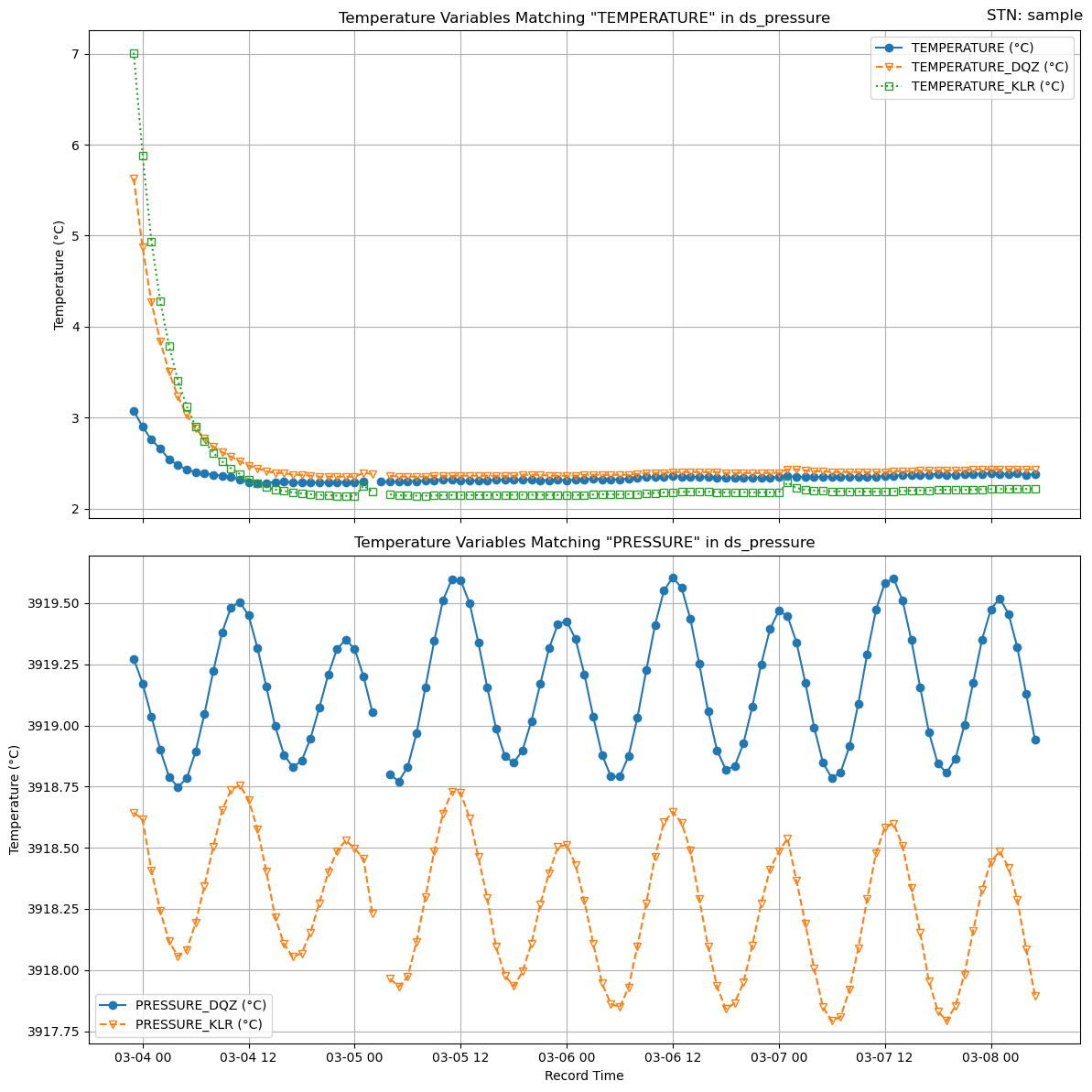
plotters.plot_temperature_variables(ds_pressure, ['TEMPERATURE', 'PRESSURE'],"sample")
print("From the analysis, we determine that PRESSURE_DQZ and PRESSURE_PIES are identical.")
From the analysis, we determine that PRESSURE_DQZ and PRESSURE_PIES are identical.
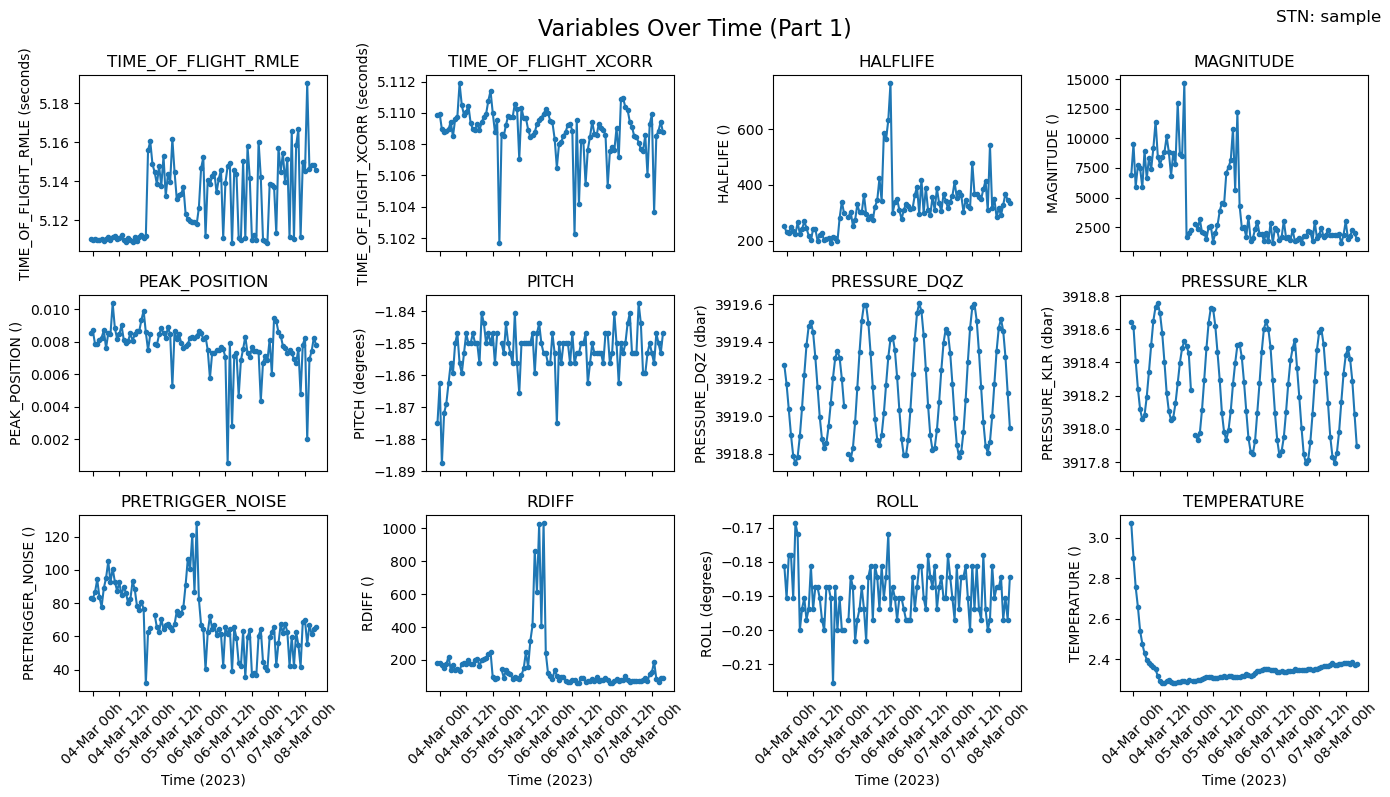
Diagnostics & basic statistics
1. Plot variables which are against time
[8]:
# Call the function to plot all variables against RECORD_TIME
#fig, axs = plotters.plot_all_variables_against_time(ds_pressure, time_var='RECORD_TIME')
fig,axs = plotters.plot_all_variables(ds_pressure, 'sample')
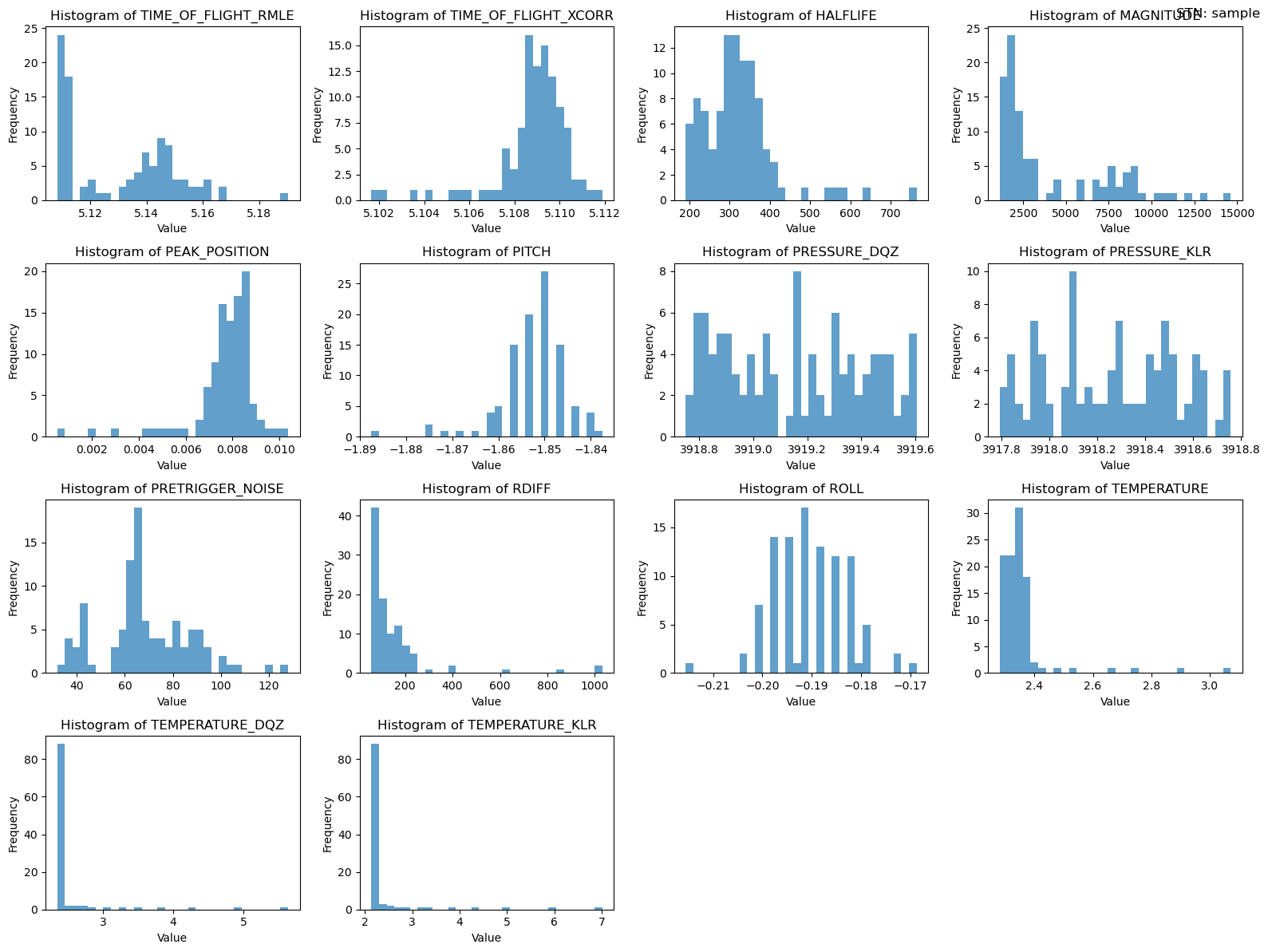
2. Histograms
[9]:
# Example usage
plotters.plot_histograms(ds_pressure, "sample")
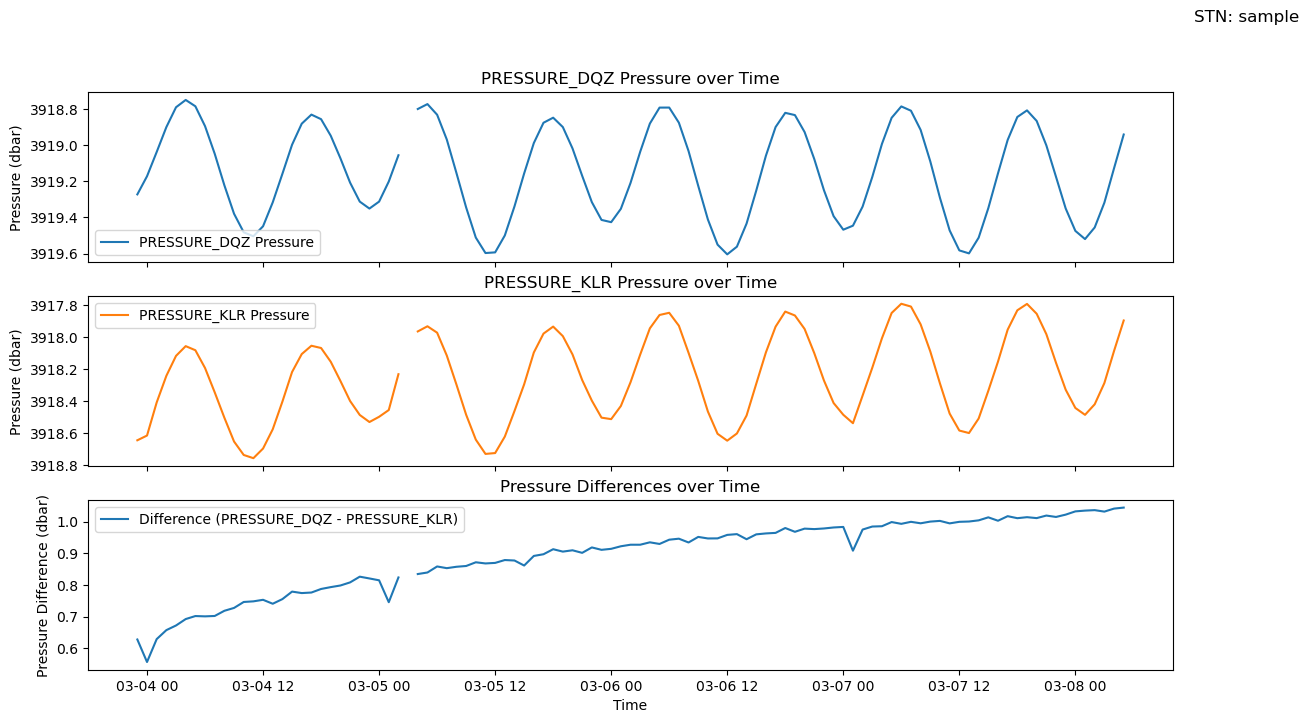
3. Compare pressure
Plot the pressure from the ambient (KLR) and transfer (DQZ)
[10]:
# Example usage
fig, axs = plotters.compare_pressure(ds_pressure, ['PRESSURE_DQZ', 'PRESSURE_KLR'], "sample")
Plot the pressure during an AZA sequence (transfer, ambient and low)
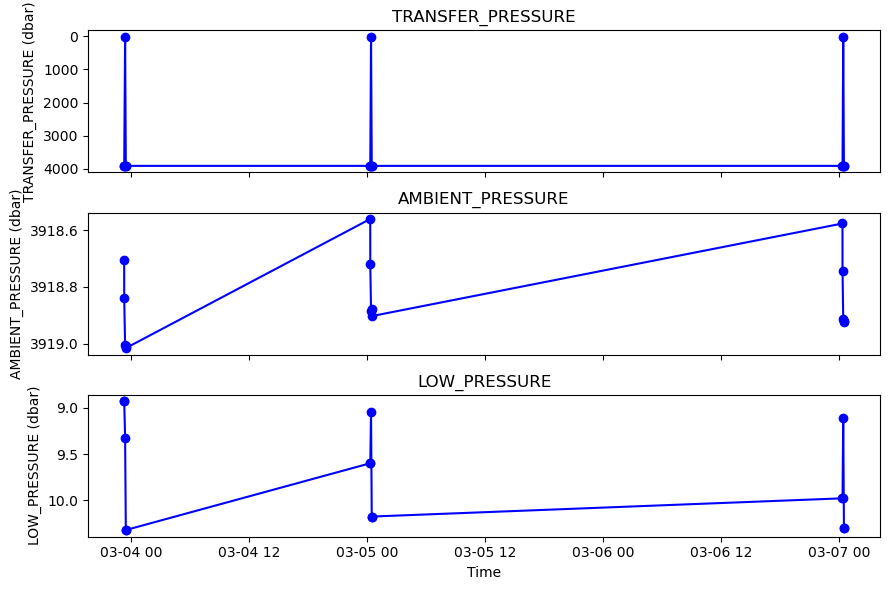
[11]:
variables_to_compare=['TRANSFER_PRESSURE','AMBIENT_PRESSURE','LOW_PRESSURE']
# Example usage
fig, axs = plotters.plot_AZA_pressure(ds_AZA, variables_to_compare)
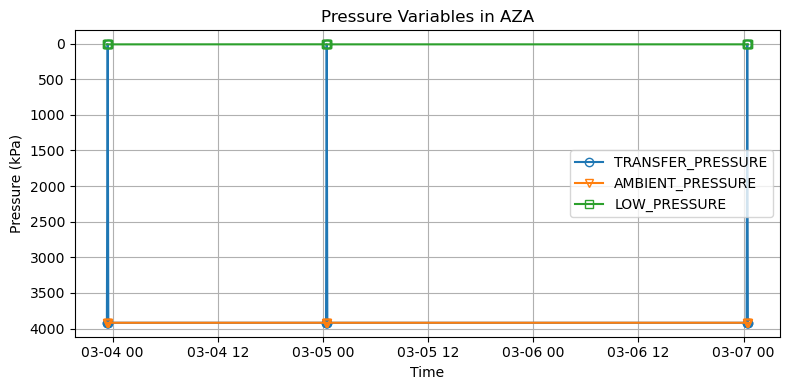
[12]:
# Plot all pressure variables in ds_AZA on the same axes with symbols and lines
plt.figure(figsize=(8, 4))
pressure_vars = ['TRANSFER_PRESSURE', 'AMBIENT_PRESSURE', 'LOW_PRESSURE']
time_var = 'SAMPLE_TIME'
# Define marker styles to cycle through
markers = ['o', 'v', 's', 'D', '^']
for idx, var in enumerate(pressure_vars):
plt.plot(
ds_AZA[time_var],
ds_AZA[var],
label=var,
marker=markers[idx % len(markers)],
linestyle='-',
markerfacecolor='none'
)
plt.title('Pressure Variables in AZA')
plt.xlabel('Time')
plt.ylabel('Pressure (kPa)')
plt.legend()
plt.grid(True)
plt.gca().invert_yaxis() # Invert the y-axis
plt.tight_layout()
plt.show()
[ ]:
[13]:
# Let's test the convertAZA function call to see what parameters are being passed
import sys
import pathlib
sys.path.append('/Users/eddifying/Cloudfree/gitlab-cloudfree/fetchAZA')
from fetchAZA import convertAZA
# Let's inspect the function signature
import inspect
sig = inspect.signature(convertAZA.convertAZA)
print("Function signature:")
print(sig)
# Let's see what the default values are
for name, param in sig.parameters.items():
print(f"{name}: default = {param.default}")
Function signature:
(data_path, fn, STN='sample', deploy_date='2000-01-01', recovery_date='2099-01-01', latitude='0', longitude='0', water_depth='0', keys=['DQZ', 'PIES', 'INC', 'TMP', 'KLR'], cleanup=True, overwrite=None)
data_path: default = <class 'inspect._empty'>
fn: default = <class 'inspect._empty'>
STN: default = sample
deploy_date: default = 2000-01-01
recovery_date: default = 2099-01-01
latitude: default = 0
longitude: default = 0
water_depth: default = 0
keys: default = ['DQZ', 'PIES', 'INC', 'TMP', 'KLR']
cleanup: default = True
overwrite: default = None
[14]:
# Let's test a simplified call to see what's happening
import os
data_path = '/Users/eddifying/Cloudfree/gitlab-cloudfree/fetchAZA/data'
fn = 'sample_data.csv'
# Test the function call that should work
print("Testing function call...")
print("This should set overwrite=True and not prompt:")
# Let's simulate the call that should not prompt
test_call = {
'data_path': data_path,
'fn': fn,
'STN': 'sample',
'deploy_date': '2023-02-27',
'recovery_date': '2023-03-08T08:00:00',
'latitude': 26.5,
'longitude': -76.75,
'water_depth': -3800,
'cleanup': True,
'overwrite': True
}
print("Parameters that will be passed:")
for key, value in test_call.items():
print(f" {key}: {value}")
Testing function call...
This should set overwrite=True and not prompt:
Parameters that will be passed:
data_path: /Users/eddifying/Cloudfree/gitlab-cloudfree/fetchAZA/data
fn: sample_data.csv
STN: sample
deploy_date: 2023-02-27
recovery_date: 2023-03-08T08:00:00
latitude: 26.5
longitude: -76.75
water_depth: -3800
cleanup: True
overwrite: True
[15]:
# Let's test the save_dataset function directly to see if it's working
import sys
sys.path.append('/Users/eddifying/Cloudfree/gitlab-cloudfree/fetchAZA')
from fetchAZA import writers
import xarray as xr
import numpy as np
import tempfile
import os
# Create a simple test datasety
test_ds = xr.Dataset({
'test_var': (['time'], np.random.rand(5))
}, coords={'time': np.arange(5)})
# Create a temporary file
with tempfile.NamedTemporaryFile(suffix='.nc', delete=False) as tmp:
tmp_file = tmp.name
# Save it once
result1 = writers.save_dataset(test_ds, tmp_file, overwrite=True)
print(f"First save result: {result1}")
print(f"File exists: {os.path.exists(tmp_file)}")
# Try to save again with overwrite=True (should not prompt)
print("\nTesting overwrite=True (should not prompt)...")
result2 = writers.save_dataset(test_ds, tmp_file, overwrite=True)
print(f"Second save result: {result2}")
# Clean up
os.remove(tmp_file)
print("Test completed")
First save result: True
File exists: True
Testing overwrite=True (should not prompt)...
Second save result: True
Test completed