Demo: Instrument-Level Processing (Compact Workflow)
This notebook walks through the complete instrument-level processing pipeline in the oceanarray codebase, from raw files to science-ready datasets. It demonstrates the same processing steps as demo_stage1.ipynb and demo_stage2.ipynb but in a more compact, streamlined format.
Processing Overview
Stage 1: Format Conversion (*_raw.nc)
Purpose: Convert raw instrument files to standardized NetCDF format
Input: Raw instrument files (
.cnv,.rsk,.dat,.mat)Output: Standardized NetCDF files (
*_raw.nc)Processing: Uses
oceanarray.stage1.MooringProcessor- same asdemo_stage1.ipynb
Stage 2: Temporal Corrections & Trimming (*_use.nc)
Purpose: Apply clock corrections and trim to deployment periods
Input: Stage1 files (
*_raw.nc) + updated YAML with clock offsetsOutput: Time-corrected files (
*_use.nc)Processing: Uses
oceanarray.stage2.process_multiple_moorings_stage2- same asdemo_stage2.ipynb
Stage 3: Calibrations & Corrections (Optional)
Purpose: Apply sensor-specific calibrations and corrections
Status: Commented out sections showing how to apply additional calibrations
Stage 4: Format Conversion (Optional)
Purpose: Convert to OceanSites or other standardized formats
Status: Commented out sections for format conversion
Key Features
Compact Format: Covers the same ground as separate stage notebooks in one place
Instrument-Level Processing: Each instrument processed independently before mooring-level coordination
Multiple Instrument Types: Handles various instrument types with analysis functions
Visualization: Includes plotting and analysis of processed results
Metadata Management: YAML configuration files drive processing parameters
Comparison with Other Notebooks
vs demo_stage1.ipynb: Same Stage1 processing but more concise
vs demo_stage2.ipynb: Same Stage2 processing but integrated workflow
vs demo_step1.ipynb: Focuses on individual instruments rather than mooring-level time gridding
Choose this notebook if you want a complete instrument processing workflow in one place, or use the separate stage notebooks for more detailed exploration of each processing step.
[1]:
from pathlib import Path
from datetime import datetime
import os
import sys
import yaml
import scipy.io
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import xarray as xr
from oceanarray.stage1 import MooringProcessor, process_multiple_moorings, stage1_mooring
from oceanarray import writers
from ctd_tools.readers import NortekAsciiReader, AdcpMatlabReader
from ctd_tools.plotters import TimeSeriesPlotter
from ctd_tools.writers import NetCdfWriter
Matplotlib is building the font cache; this may take a moment.
/home/runner/micromamba/envs/TEST/lib/python3.13/site-packages/pycnv/pycnv.py:7: UserWarning: pkg_resources is deprecated as an API. See https://setuptools.pypa.io/en/latest/pkg_resources.html. The pkg_resources package is slated for removal as early as 2025-11-30. Refrain from using this package or pin to Setuptools<81.
import pkg_resources
Configuration
Set up the base directory and mooring lists for processing
[2]:
# Base directory containing the mooring data
basedir = '../data/'
# Define mooring lists
single_test = ['dsE_1_2018']
# Choose which set to process
moorlist = single_test
print(f"Base directory: {basedir}")
print(f"Processing {len(moorlist)} moorings: {moorlist}")
Base directory: ../data/
Processing 1 moorings: ['dsE_1_2018']
Stage 1: Load raw instrument files and convert to *_raw.nc
[3]:
# Initialize the processor
processor = MooringProcessor(basedir)
# Process each mooring individually with detailed output
results = {}
for mooring_name in moorlist:
print(f"\n{'='*60}")
print(f"Processing mooring: {mooring_name}")
print(f"{'='*60}")
success = processor.process_mooring(mooring_name)
results[mooring_name] = success
status = "✅ SUCCESS" if success else "❌ FAILED"
print(f"\nResult for {mooring_name}: {status}")
# Print final summary
print(f"\n{'='*60}")
print("FINAL PROCESSING SUMMARY")
print(f"{'='*60}")
successful = sum(results.values())
total = len(results)
print(f"Successfully processed: {successful}/{total} moorings")
for mooring, success in results.items():
status = "✅" if success else "❌"
print(f"{status} {mooring}")
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606363_2018_08_27.cnv
CRITICAL:pycnv:Could not open file:../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606363_2018_08_27.cnv (Exception: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606363_2018_08_27.cnv')
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe16/DSE18_sbe16_2419efw1.cnv
WARNING:pycnv:Could not parse custom header ** recovery DS-E-18
WARNING:pycnv:Could not compute datetime dates based on timeM
INFO:pycnv:Dates computed based on timeS
============================================================
Processing mooring: dsE_1_2018
============================================================
Processing mooring: dsE_1_2018
EXCEPT: Error reading file moor/raw/msm76_2018/sbe56/DSE18_SBE05606363_2018_08_27.cnv: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606363_2018_08_27.cnv'
Date
Computing date
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606401_2018_08_27.cnv
CRITICAL:pycnv:Could not open file:../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606401_2018_08_27.cnv (Exception: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606401_2018_08_27.cnv')
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606402_2018_08_27.cnv
CRITICAL:pycnv:Could not open file:../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606402_2018_08_27.cnv (Exception: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606402_2018_08_27.cnv')
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05608482_2018_08_27.cnv
WARNING:pycnv:Could not parse custom header * ** recovery DS-E-18
WARNING:pycnv:parse_header() start_time: Could not decode time: ( Aug 11 2018 12:00:00 )unconverted data remains: locale('C', 'UTF-8')
WARNING:pycnv:parse_header() start_time: Could not decode time: ( Aug 11 2018 12:00:00 )unconverted data remains: locale(None, None)
EXCEPT: Error reading file moor/raw/msm76_2018/sbe16/DSE18_sbe16_2419efw1.cnv: Variable 'depth': Could not convert tuple of form (dims, data[, attrs, encoding]): (['time'], None) to Variable.
EXCEPT: Error reading file moor/raw/msm76_2018/sbe56/DSE18_SBE05606401_2018_08_27.cnv: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606401_2018_08_27.cnv'
EXCEPT: Error reading file moor/raw/msm76_2018/sbe56/DSE18_SBE05606402_2018_08_27.cnv: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606402_2018_08_27.cnv'
WARNING:pycnv:Could not compute datetime dates based on timeM
WARNING:pycnv:Could not compute datetime dates based on timeS
WARNING:pycnv:Could not compute datetime dates based on start_date and time_interval
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606365_2018_08_27.cnv
CRITICAL:pycnv:Could not open file:../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606365_2018_08_27.cnv (Exception: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606365_2018_08_27.cnv')
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606409_2018_08_27.cnv
CRITICAL:pycnv:Could not open file:../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606409_2018_08_27.cnv (Exception: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606409_2018_08_27.cnv')
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606397_2018_08_27.cnv
CRITICAL:pycnv:Could not open file:../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606397_2018_08_27.cnv (Exception: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606397_2018_08_27.cnv')
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606366_2018_08_27.cnv
CRITICAL:pycnv:Could not open file:../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606366_2018_08_27.cnv (Exception: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606366_2018_08_27.cnv')
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606394_2018_08_27.cnv
CRITICAL:pycnv:Could not open file:../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606394_2018_08_27.cnv (Exception: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606394_2018_08_27.cnv')
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606370_2018_08_27.cnv
CRITICAL:pycnv:Could not open file:../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606370_2018_08_27.cnv (Exception: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606370_2018_08_27.cnv')
Date
Computing date
EXCEPT: Error reading file moor/raw/msm76_2018/sbe56/DSE18_SBE05608482_2018_08_27.cnv: 'NoneType' object has no attribute 'strftime'
EXCEPT: Error reading file moor/raw/msm76_2018/sbe56/DSE18_SBE05606365_2018_08_27.cnv: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606365_2018_08_27.cnv'
EXCEPT: Error reading file moor/raw/msm76_2018/sbe56/DSE18_SBE05606409_2018_08_27.cnv: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606409_2018_08_27.cnv'
EXCEPT: Error reading file moor/raw/msm76_2018/sbe56/DSE18_SBE05606397_2018_08_27.cnv: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606397_2018_08_27.cnv'
EXCEPT: Error reading file moor/raw/msm76_2018/sbe56/DSE18_SBE05606366_2018_08_27.cnv: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606366_2018_08_27.cnv'
EXCEPT: Error reading file moor/raw/msm76_2018/sbe56/DSE18_SBE05606394_2018_08_27.cnv: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606394_2018_08_27.cnv'
EXCEPT: Error reading file moor/raw/msm76_2018/sbe56/DSE18_SBE05606370_2018_08_27.cnv: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606370_2018_08_27.cnv'
EXCEPT: Error reading file moor/raw/msm76_2018/sbe16/DSE18_sbe16_2418.hex: Unknown file type: sbe-hex
EXCEPT: Error reading file moor/raw/msm76_2018/tr1050/DSE18_013889_20180827_1349.mat: Unknown file type: rbr-matlab
--> Processing rbrsolo: moor/raw/msm76_2018/rbrsolo/DSE18_101651_20180827_1541.rsk
Creating output file: moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101651_raw.nc
ERROR: Failed to process moor/raw/msm76_2018/rbrsolo/DSE18_101651_20180827_1541.rsk: Invalid value for attr 'processor_input_file_type': <function RbrRskLegacyReader.format_name at 0x7fab57442020>. For serialization to netCDF files, its value must be of one of the following types: str, Number, ndarray, number, list, tuple, bytes
EXCEPT: Error reading file moor/raw/msm76_2018/tr1050/DSE18_015580_20180827_1403.mat: Unknown file type: rbr-matlab
--> Processing rbrsolo: moor/raw/msm76_2018/rbrsolo/DSE18_101647_20180827_1551.rsk
Creating output file: moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101647_raw.nc
ERROR: Failed to process moor/raw/msm76_2018/rbrsolo/DSE18_101647_20180827_1551.rsk: Invalid value for attr 'processor_input_file_type': <function RbrRskLegacyReader.format_name at 0x7fab57442020>. For serialization to netCDF files, its value must be of one of the following types: str, Number, ndarray, number, list, tuple, bytes
EXCEPT: Error reading file moor/raw/msm76_2018/tr1050/DSE18_013874_20180827_1410.mat: Unknown file type: rbr-matlab
--> Processing rbrsolo: moor/raw/msm76_2018/rbrsolo/DSE18_101645_20180827_1553.rsk
Creating output file: moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101645_raw.nc
ERROR: Failed to process moor/raw/msm76_2018/rbrsolo/DSE18_101645_20180827_1553.rsk: Invalid value for attr 'processor_input_file_type': <function RbrRskLegacyReader.format_name at 0x7fab57442020>. For serialization to netCDF files, its value must be of one of the following types: str, Number, ndarray, number, list, tuple, bytes
EXCEPT: Error reading file moor/raw/msm76_2018/tr1050/DSE18_015574_20180827_1407.mat: Unknown file type: rbr-matlab
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/microcat/DSE18_SBE37SM_RS232_03707518_2018_08_26efw1.cnv
--> Processing rbrsolo: moor/raw/msm76_2018/rbrsolo/DSE18_101646_20180827_1548.rsk
Creating output file: moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101646_raw.nc
ERROR: Failed to process moor/raw/msm76_2018/rbrsolo/DSE18_101646_20180827_1548.rsk: Invalid value for attr 'processor_input_file_type': <function RbrRskLegacyReader.format_name at 0x7fab57442020>. For serialization to netCDF files, its value must be of one of the following types: str, Number, ndarray, number, list, tuple, bytes
EXCEPT: Error reading file moor/raw/msm76_2018/tr1050/DSE18_015577_20180827_1338.mat: Unknown file type: rbr-matlab
WARNING:pycnv:Could not compute datetime dates based on timeM
WARNING:pycnv:Could not compute datetime dates based on timeS
INFO:pycnv:Dates computed based on start_date and time_interval
Date
Computing date
INFO:pycnv: Opening file: ../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606364_2018_08_27.cnv
CRITICAL:pycnv:Could not open file:../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606364_2018_08_27.cnv (Exception: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606364_2018_08_27.cnv')
--> Processing microcat: moor/raw/msm76_2018/microcat/DSE18_SBE37SM_RS232_03707518_2018_08_26efw1.cnv
Creating output file: moor/proc/dsE_1_2018/microcat/dsE_1_2018_7518_raw.nc
Removing variable: potential_temperature
Removing variable: julian_days_offset
Removing variable: density
Removing coordinate: depth
Removing coordinate: latitude
Removing coordinate: longitude
EXCEPT: Error reading file moor/raw/msm76_2018/sbe56/DSE18_SBE05606364_2018_08_27.cnv: [Errno 2] No such file or directory: '../data/moor/raw/msm76_2018/sbe56/DSE18_SBE05606364_2018_08_27.cnv'
Completed processing: 1/23 instruments successful
Result for dsE_1_2018: ✅ SUCCESS
============================================================
FINAL PROCESSING SUMMARY
============================================================
Successfully processed: 1/1 moorings
✅ dsE_1_2018
[4]:
# Analyze processed files
def analyze_processed_mooring(mooring_name, basedir, instrument_type='microcat'):
"""Analyze processed files for a specific mooring and instrument type."""
proc_dir = Path(basedir) / 'moor/proc' / mooring_name / instrument_type
fig_dir = proc_dir
if not proc_dir.exists():
print(f"Directory not found: {proc_dir}")
return
print(f"Analyzing processed files in: {proc_dir}")
# Find all NetCDF files
nc_files = list(proc_dir.glob('*raw.nc'))
if not nc_files:
print("No NetCDF files found")
return
print(f"Found {len(nc_files)} NetCDF files:")
for file_path in nc_files:
try:
print(f"\n📄 Loading: {file_path.name}")
with xr.open_dataset(file_path) as dataset:
print(f" Variables: {list(dataset.data_vars)}")
print(f" Attributes: {list(dataset.attrs.keys())}")
# Extract key metadata
mooring_name_attr = dataset.attrs.get('mooring_name', 'Unknown')
instrument_name = dataset.get('instrument', 'Unknown').values if 'instrument' in dataset else 'Unknown'
serial_number = dataset.get('serial_number', 0).values if 'serial_number' in dataset else 0
print(f" Mooring: {mooring_name_attr}")
print(f" Instrument: {instrument_name}")
print(f" Serial: {serial_number}")
# Plot temperature if available
if 'temperature' in dataset.data_vars:
fig, ax = plt.subplots(figsize=(12, 4))
ax.plot(dataset['time'], dataset['temperature'], 'b-', linewidth=0.5)
ax.set_ylabel('Temperature (°C)')
ax.set_title(f'{mooring_name_attr}: {instrument_name} {serial_number} - Temperature')
ax.set_xlabel('Time')
ax.grid(True, alpha=0.3)
# Add stats
temp_mean = dataset['temperature'].mean().values
temp_std = dataset['temperature'].std().values
ax.text(0.02, 0.98, f'Mean: {temp_mean:.2f}°C\nStd: {temp_std:.2f}°C',
transform=ax.transAxes, verticalalignment='top',
bbox=dict(boxstyle='round', facecolor='white', alpha=0.8))
plt.tight_layout()
plt.show()
# Save plot
plot_name = f"{mooring_name_attr}_{instrument_name}_{serial_number}_temperature_raw.png"
fig.savefig(fig_dir / plot_name, dpi=150, bbox_inches='tight')
print(f" 📊 Plot saved: {plot_name}")
except Exception as e:
print(f" ❌ Error loading {file_path.name}: {e}")
# Example usage
if results and any(results.values()):
# Analyze the first successfully processed mooring
successful_mooring = next(mooring for mooring, success in results.items() if success)
print(f"\nAnalyzing successful mooring: {successful_mooring}")
analyze_processed_mooring(successful_mooring, basedir, 'microcat')
analyze_processed_mooring(successful_mooring, basedir, 'sbe16')
analyze_processed_mooring(successful_mooring, basedir, 'sbe56')
else:
print("No successfully processed moorings to analyze")
Analyzing successful mooring: dsE_1_2018
Analyzing processed files in: ../data/moor/proc/dsE_1_2018/microcat
Found 1 NetCDF files:
📄 Loading: dsE_1_2018_7518_raw.nc
Variables: ['temperature', 'salinity', 'conductivity', 'pressure', 'serial_number', 'InstrDepth', 'instrument', 'clock_offset', 'start_time', 'end_time']
Attributes: ['latitude', 'longitude', 'CreateTime', 'DataType', 'mooring_name', 'waterdepth', 'deployment_latitude', 'deployment_longitude', 'deployment_time', 'seabed_latitude', 'seabed_longitude', 'recovery_time']
Mooring: dsE_1_2018
Instrument: microcat
Serial: 7518
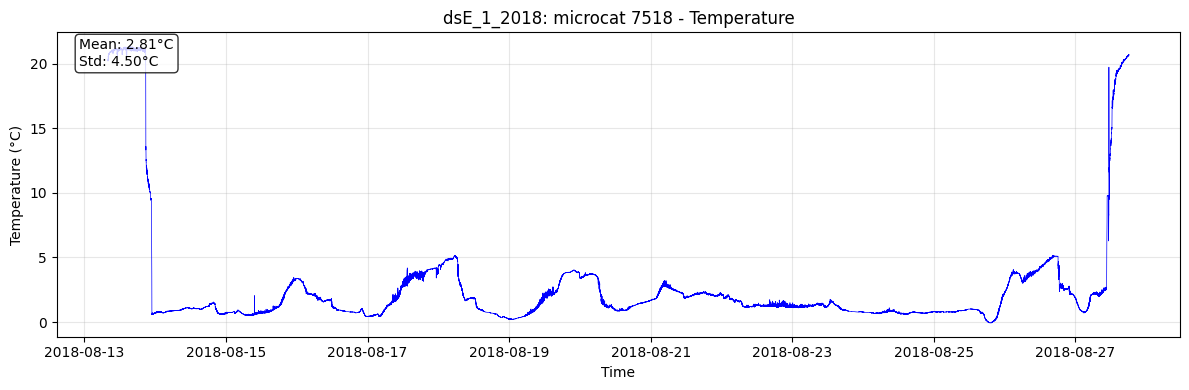
📊 Plot saved: dsE_1_2018_microcat_7518_temperature_raw.png
Analyzing processed files in: ../data/moor/proc/dsE_1_2018/sbe16
No NetCDF files found
Analyzing processed files in: ../data/moor/proc/dsE_1_2018/sbe56
No NetCDF files found
Stage 2: Trim to deployment period
[5]:
from oceanarray.stage2 import Stage2Processor, process_multiple_moorings_stage2
results = process_multiple_moorings_stage2(moorlist, basedir)
==================================================
Processing Stage 2 for mooring dsE_1_2018
==================================================
Starting Stage 2 processing for mooring: dsE_1_2018
Deployment time: 2018-08-12T22:44:00.000000000
Recovery time: 2018-08-26T10:38:00.000000000
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6363_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe16/dsE_1_2018_2419_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6401_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6402_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_8482_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6365_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6409_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6397_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6366_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6394_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6370_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe16/dsE_1_2018_2418_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/tr1050/dsE_1_2018_13889_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101651_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/tr1050/dsE_1_2018_15580_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101647_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/tr1050/dsE_1_2018_13874_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101645_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/tr1050/dsE_1_2018_15574_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101646_raw.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/tr1050/dsE_1_2018_15577_raw.nc
Processing microcat serial 7518
Applying clock offset: -2050 seconds
Trimming start to deployment time: 2018-08-12T22:44:00.000000000
Trimming end to recovery time: 2018-08-26T10:38:00.000000000
Trimmed from 124619 to 113473 records
Final time range: 2018-08-13T07:25:51.008000000 to 2018-08-26T10:37:50.979200000
Successfully wrote: ../data/moor/proc/dsE_1_2018/microcat/dsE_1_2018_7518_use.nc
WARNING: Raw file not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6364_raw.nc
Stage 2 completed: 1/23 instruments successful
Stage 3: Apply calibrations + corrections
[6]:
#ds_cal = process_rodb.apply_microcat_calibration_from_txt(data_dir / 'wb1_12_2015_005.microcat.txt', data_dir / 'wb1_12_2015_6123.use')
#ds_cal
[7]:
#fig = plotters.plot_microcat(ds_cal)
[8]:
#ds_diff = tools.calc_ds_difference(ds_cal, ds2)
#fig = plotters.plot_microcat(ds_diff)
#fig.suptitle("difference between *.use and *.microcat")
Stage 4: Convert to OceanSites format
[9]:
if 0:
metadata_txt = data_dir / 'wb1_12_2015_6123.use'
config_dir = Path("..") / "oceanarray" / "config"
var_map_yaml = config_dir / "OS1_var_names.yaml"
vocab_yaml = config_dir / "OS1_vocab_attrs.yaml"
sensor_yaml = config_dir / "OS1_sensor_attrs.yaml"
project_yaml = config_dir / "project_RAPID.yaml"
ds_OS = convertOS.convert_rodb_to_oceansites(ds_cal, metadata_txt, var_map_yaml, vocab_yaml, sensor_yaml=sensor_yaml,project_yaml=project_yaml)
ds_OS
[10]:
if 0:
filepath = writers.save_OS_instrument(ds_OS, basedir)
print(filepath)