Demo: mooring-level processing (Step 1)
This notebook demonstrates the first of the mooring-level processing steps, time_gridding.
Step 1: Time Gridding and Optional Filtering Demo
This notebook demonstrates the Step 1 processing workflow for mooring data:
Loading multiple instrument datasets
Optional time-domain filtering (applied BEFORE interpolation)
Interpolating onto a common time grid
Combining into a unified mooring dataset
Key Point: Filtering is applied to individual instrument records on their native time grids BEFORE interpolation to preserve data integrity.
[1]:
import numpy as np
import pandas as pd
import xarray as xr
import matplotlib.pyplot as plt
from pathlib import Path
import yaml
# Import the time gridding module
from oceanarray.time_gridding import (
TimeGriddingProcessor,
time_gridding_mooring,
process_multiple_moorings_time_gridding
)
# Set up plotting
plt.style.use('default')
Configuration
First, let’s set up our data paths and examine the mooring configuration.
[2]:
# Set your data paths here
basedir = '../data'
mooring_name = 'dsE_1_2018'
# Construct paths
proc_dir = Path(basedir) / 'moor' / 'proc' / mooring_name
config_file = proc_dir / f"{mooring_name}.mooring.yaml"
print(f"Processing directory: {proc_dir}")
print(f"Configuration file: {config_file}")
print(f"Config exists: {config_file.exists()}")
Processing directory: ../data/moor/proc/dsE_1_2018
Configuration file: ../data/moor/proc/dsE_1_2018/dsE_1_2018.mooring.yaml
Config exists: True
[3]:
# Load and examine the mooring configuration
if config_file.exists():
with open(config_file, 'r') as f:
config = yaml.safe_load(f)
print("Mooring Configuration:")
print(f"Name: {config['name']}")
print(f"Water depth: {config.get('waterdepth', 'unknown')} m")
print(f"Location: {config.get('latitude', 'unknown')}°N, {config.get('longitude', 'unknown')}°E")
print(f"\nInstruments ({len(config.get('instruments', []))}):")
for i, inst in enumerate(config.get('instruments', [])):
print(f" {i+1}. {inst.get('instrument', 'unknown')} "
f"(serial: {inst.get('serial num.', 'unknown')}) at {inst.get('depth', 'unknown')} m")
else:
print("Configuration file not found!")
print("Please check your data path and mooring name.")
Mooring Configuration:
Name: dsE_1_2018
Water depth: 929 m
Location: 65.47561666666667°N, -29.570083333333333°E
Instruments (23):
1. sbe56 (serial: unknown) at 629 m
2. sbe16 (serial: unknown) at 679 m
3. sbe56 (serial: unknown) at 689 m
4. sbe56 (serial: unknown) at 699 m
5. sbe56 (serial: unknown) at 709 m
6. sbe56 (serial: unknown) at 719 m
7. sbe56 (serial: unknown) at 729 m
8. sbe56 (serial: unknown) at 739 m
9. sbe56 (serial: unknown) at 749 m
10. sbe56 (serial: unknown) at 759 m
11. sbe56 (serial: unknown) at 769 m
12. sbe16 (serial: unknown) at 780 m
13. tr1050 (serial: unknown) at 790 m
14. rbrsolo (serial: unknown) at 800 m
15. tr1050 (serial: unknown) at 810 m
16. rbrsolo (serial: unknown) at 820 m
17. tr1050 (serial: unknown) at 830 m
18. rbrsolo (serial: unknown) at 840 m
19. tr1050 (serial: unknown) at 850 m
20. rbrsolo (serial: unknown) at 860 m
21. tr1050 (serial: unknown) at 870 m
22. microcat (serial: unknown) at 880 m
23. sbe56 (serial: unknown) at 905 m
Examine individual instrument files
Let’s look at the individual instrument files before processing to understand the different sampling rates and data characteristics.
[4]:
# Find and examine individual instrument files
file_suffix = "_use"
instrument_files = []
instrument_datasets = []
rows = []
if config_file.exists():
for inst_config in config.get("instruments", []):
instrument_type = inst_config.get("instrument", "unknown")
serial = inst_config.get("serial", 0)
depth = inst_config.get("depth", 0)
# Look for the file
filename = f"{mooring_name}_{serial}{file_suffix}.nc"
filepath = proc_dir / instrument_type / filename
if filepath.exists():
ds = xr.open_dataset(filepath)
instrument_files.append(filepath)
instrument_datasets.append(ds)
# Time coverage
t0, t1 = ds.time.values[0], ds.time.values[-1]
npoints = len(ds.time)
# Median sampling interval
time_diff = np.diff(ds.time.values) / np.timedelta64(1, "m") # in minutes
median_interval = np.nanmedian(time_diff)
if median_interval > 1:
sampling = f"{median_interval:.1f} min"
else:
sampling = f"{median_interval*60:.1f} sec"
# Collect a row for the table
rows.append(
{
"Instrument": instrument_type,
"Serial": serial,
"Depth [m]": depth,
"File": filepath.name,
"Start": str(t0)[:19],
"End": str(t1)[:19],
"Points": npoints,
"Sampling": sampling,
"Variables": ", ".join(list(ds.data_vars)),
}
)
else:
rows.append(
{
"Instrument": instrument_type,
"Serial": serial,
"Depth [m]": depth,
"File": "MISSING",
"Start": "",
"End": "",
"Points": 0,
"Sampling": "",
"Variables": "",
}
)
# Make a DataFrame summary
summary = pd.DataFrame(rows)
pd.set_option("display.max_colwidth", 80) # allow long var lists
print(summary.to_markdown(index=False))
print(f"\nFound {len(instrument_datasets)} instrument datasets")
| Instrument | Serial | Depth [m] | File | Start | End | Points | Sampling | Variables |
|:-------------|---------:|------------:|:-----------------------|:--------------------|:--------------------|---------:|:-----------|:-------------------------------------------------------------------------------------------------------------------------|
| sbe56 | 6363 | 629 | MISSING | | | 0 | | |
| sbe16 | 2419 | 679 | MISSING | | | 0 | | |
| sbe56 | 6401 | 689 | MISSING | | | 0 | | |
| sbe56 | 6402 | 699 | MISSING | | | 0 | | |
| sbe56 | 8482 | 709 | MISSING | | | 0 | | |
| sbe56 | 6365 | 719 | MISSING | | | 0 | | |
| sbe56 | 6409 | 729 | MISSING | | | 0 | | |
| sbe56 | 6397 | 739 | MISSING | | | 0 | | |
| sbe56 | 6366 | 749 | MISSING | | | 0 | | |
| sbe56 | 6394 | 759 | MISSING | | | 0 | | |
| sbe56 | 6370 | 769 | MISSING | | | 0 | | |
| sbe16 | 2418 | 780 | MISSING | | | 0 | | |
| tr1050 | 13889 | 790 | MISSING | | | 0 | | |
| rbrsolo | 101651 | 800 | MISSING | | | 0 | | |
| tr1050 | 15580 | 810 | MISSING | | | 0 | | |
| rbrsolo | 101647 | 820 | MISSING | | | 0 | | |
| tr1050 | 13874 | 830 | MISSING | | | 0 | | |
| rbrsolo | 101645 | 840 | MISSING | | | 0 | | |
| tr1050 | 15574 | 850 | MISSING | | | 0 | | |
| rbrsolo | 101646 | 860 | MISSING | | | 0 | | |
| tr1050 | 15577 | 870 | MISSING | | | 0 | | |
| microcat | 7518 | 880 | dsE_1_2018_7518_use.nc | 2018-08-13T07:25:51 | 2018-08-26T10:37:50 | 113473 | 10.0 sec | temperature, salinity, conductivity, pressure, serial_number, InstrDepth, instrument, clock_offset, start_time, end_time |
| sbe56 | 6364 | 905 | MISSING | | | 0 | | |
Found 1 instrument datasets
Process with time gridding (no filtering)
[5]:
# Process without filtering
print("Processing mooring with time gridding only (no filtering)...")
print("="*60)
result = time_gridding_mooring(mooring_name, basedir, file_suffix='_use')
print(f"\nProcessing result: {'SUCCESS' if result else 'FAILED'}")
Processing mooring with time gridding only (no filtering)...
============================================================
Starting Step 1 (time gridding) processing for mooring: dsE_1_2018
Using files with suffix: _use
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6363_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe16/dsE_1_2018_2419_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6401_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6402_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_8482_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6365_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6409_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6397_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6366_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6394_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6370_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe16/dsE_1_2018_2418_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/tr1050/dsE_1_2018_13889_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101651_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/tr1050/dsE_1_2018_15580_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101647_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/tr1050/dsE_1_2018_13874_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101645_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/tr1050/dsE_1_2018_15574_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/rbrsolo/dsE_1_2018_101646_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/tr1050/dsE_1_2018_15577_use.nc
Loading microcat serial 7518: dsE_1_2018_7518_use.nc
WARNING: File not found: ../data/moor/proc/dsE_1_2018/sbe56/dsE_1_2018_6364_use.nc
WARNING: Missing instruments compared to YAML configuration:
- sbe56:6363
- sbe16:2419
- sbe56:6401
- sbe56:6402
- sbe56:8482
- sbe56:6365
- sbe56:6409
- sbe56:6397
- sbe56:6366
- sbe56:6394
- sbe56:6370
- sbe16:2418
- tr1050:13889
- rbrsolo:101651
- tr1050:15580
- rbrsolo:101647
- tr1050:13874
- rbrsolo:101645
- tr1050:15574
- rbrsolo:101646
- tr1050:15577
- sbe56:6364
Expected 23, found 1
Loaded 1 instrument datasets
Dataset 0 depth 880 [microcat:7518]:
Start: 2018-08-13T07:25:51, End: 2018-08-26T10:37:50
Time interval - Median: 10.02 sec, Range: 9.94 sec to 10.02 sec, Std: 0.04 sec
Variables: ['temperature', 'salinity', 'conductivity', 'pressure', 'serial_number', 'InstrDepth', 'instrument', 'clock_offset', 'start_time', 'end_time']
TIMING ANALYSIS:
Overall median interval: 0.17 min
Range of median intervals: 0.17 to 0.17 min
Common grid will use 0.17 min intervals
microcat:7518: 0.17 min -> 0.17 min (0.0% change) MINIMAL CHANGE
Common time grid: 113472 points from 2018-08-13T07:25:51.008000000 to 2018-08-26T10:37:41.008000000
INTERPOLATING FILTERED DATASETS ONTO COMMON GRID:
Successfully interpolated dataset 0
Variable 'u_velocity' not found in any dataset, skipping
Variable 'v_velocity' not found in any dataset, skipping
Successfully wrote time-gridded dataset: ../data/moor/proc/dsE_1_2018/dsE_1_2018_mooring_use.nc
Combined dataset shape: {'time': 113472, 'N_LEVELS': 1}
Variables: ['temperature', 'salinity', 'conductivity', 'pressure', 'instrument_id']
Processing result: SUCCESS
[6]:
# Load and examine the combined dataset
output_file = proc_dir / f"{mooring_name}_mooring_use.nc"
if output_file.exists():
print(f"Output file exists: {output_file}")
# Load the combined dataset
combined_ds = xr.open_dataset(output_file)
else:
print("Output file not found - processing may have failed")
Output file exists: ../data/moor/proc/dsE_1_2018/dsE_1_2018_mooring_use.nc
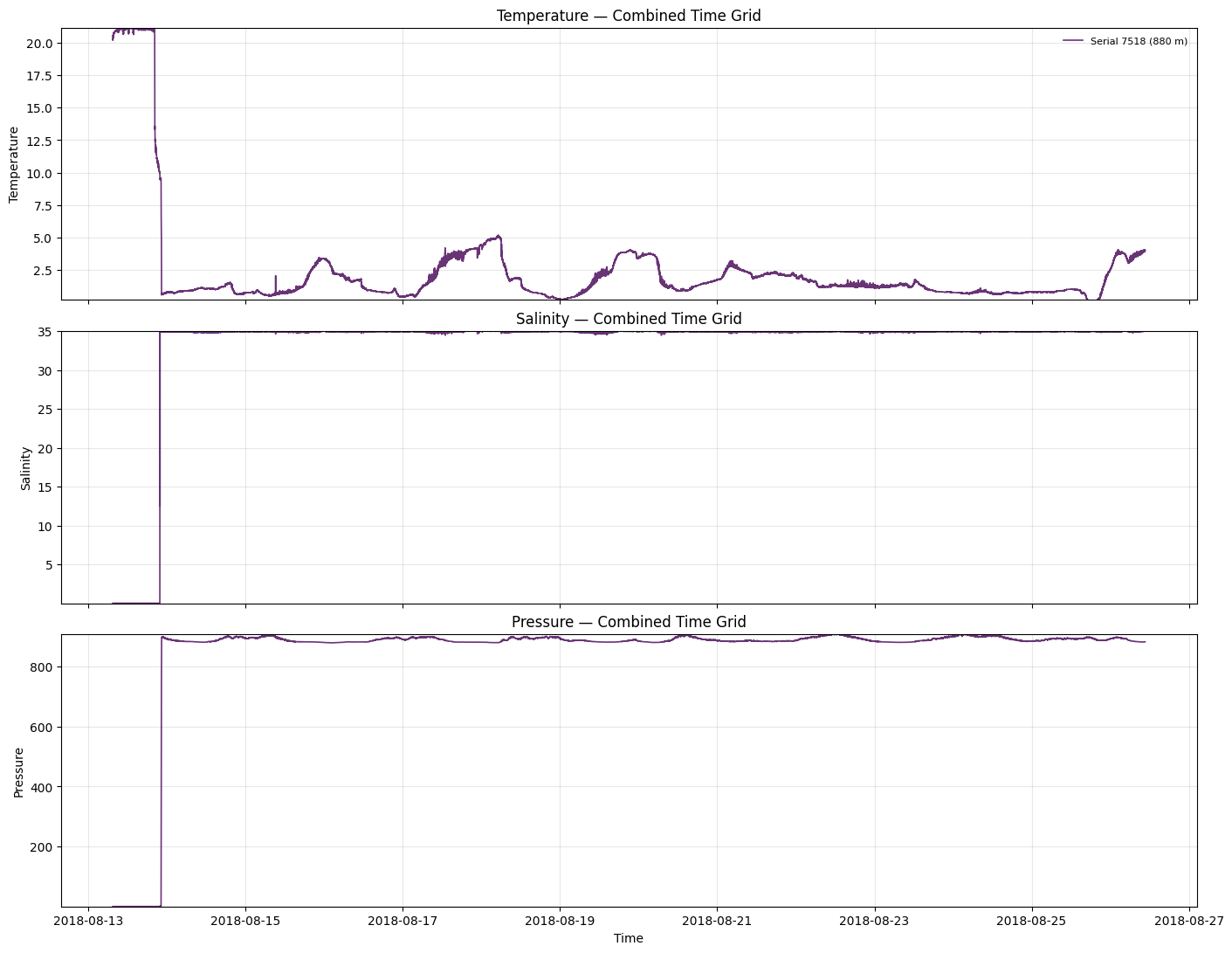
Visualize Combined Dataset
Let’s plot the combined dataset to see how the different instruments look on the common time grid.
[7]:
import numpy as np
import matplotlib.pyplot as plt
def plot_combined_timeseries(
combined_ds,
variables=("temperature", "salinity", "pressure"),
cmap_name="viridis",
line_alpha=0.8,
line_width=1.2,
percentile_limits=(1, 99),
):
"""
Plot selected variables from a combined mooring dataset as stacked time series.
Parameters
----------
combined_ds : xarray.Dataset
Must have dims: time, N_LEVELS. Optional coords: nominal_depth, serial_number.
variables : iterable[str]
Variable names to try to plot (if present in dataset).
cmap_name : str
Matplotlib colormap name for coloring by instrument level.
line_alpha : float
Line transparency.
line_width : float
Line width.
percentile_limits : (low, high)
Percentiles to use for automatic y-limits (e.g., (1, 99)).
"""
if combined_ds is None:
print("Combined dataset not available.")
return None, None
n_levels = combined_ds.sizes.get("N_LEVELS")
if n_levels is None:
raise ValueError("Dataset must contain dimension 'N_LEVELS'.")
available = [v for v in variables if v in combined_ds.data_vars]
if not available:
print("No requested variables found to plot.")
return None, None
# Colors by level
cmap = plt.get_cmap(cmap_name)
colors = cmap(np.linspace(0, 1, n_levels))
fig, axes = plt.subplots(
len(available), 1, figsize=(14, 3.6 * len(available)), sharex=True, constrained_layout=True
)
if len(available) == 1:
axes = [axes]
depth_arr = combined_ds.get("nominal_depth")
serial_arr = combined_ds.get("serial_number")
first_axis = True
for ax, var in zip(axes, available):
values_for_limits = []
for level in range(n_levels):
depth = None if depth_arr is None else depth_arr.values[level]
serial = None if serial_arr is None else serial_arr.values[level]
label = None
if first_axis:
if depth is not None and np.isfinite(depth):
label = f"Serial {serial} ({int(depth)} m)" if serial is not None else f"({int(depth)} m)"
elif serial is not None:
label = f"Serial {serial}"
da = combined_ds[var].isel(N_LEVELS=level)
da = da.where(np.isfinite(da), drop=True)
if da.size == 0:
continue
values_for_limits.append(da.values)
ax.plot(
da["time"].values,
da.values,
color=colors[level],
alpha=line_alpha,
linewidth=line_width,
label=label,
)
# Set labels and grid
ax.set_ylabel(var.replace("_", " ").title())
ax.grid(True, alpha=0.3)
ax.set_title(f"{var.replace('_', ' ').title()} — Combined Time Grid")
# Legend only once
if first_axis:
ax.legend(ncol=3, fontsize=8, loc="upper right", frameon=False)
first_axis = False
# Auto y-limits based on percentiles
if values_for_limits:
flat = np.concatenate(values_for_limits)
low, high = np.nanpercentile(flat, percentile_limits)
ax.set_ylim(low, high)
axes[-1].set_xlabel("Time")
return fig, axes
# Usage:
if 'combined_ds' in locals():
plot_combined_timeseries(combined_ds)